Introduction to R and RStudio
Last updated on 2024-12-10 | Edit this page
Overview
Questions
- How can I find my way around RStudio?
- How can I manage my projects in RStudio?
- How to interact with R for data analysis and coding?
Objectives
- Describe the purpose and functionality of each pane in RStudio
- Locate key buttons and options within the RStudio interface
- Create and manage self-contained projects in RStudio
- Define and work with variables
- Assign data to a variable
- Use mathematical and comparison operators
- Call and work with functions
Motivation
Science is a multi-step process: once you’ve designed an experiment and collected data, the real fun begins with analysis!
Although we could use a spreadsheet in Microsoft Excel or Google sheets to analyze our data, these tools are limited in their flexibility and accessibility. Critically, they also are difficult to share steps which explore and change the raw data, which is key to “reproducible” research.
Therefore, this lesson will teach you how to begin exploring your data using R and RStudio. The R program is available for Windows, Mac, and Linux operating systems, and is a freely-available where you downloaded it above. To run R, all you need is the R program.
However, to make using R easier, we will use the program RStudio, which we also downloaded above. RStudio is a free, open-source, Integrated Development Environment (IDE). It provides a built-in editor, works on all platforms (including on servers) and provides many advantages such as integration with version control and project management.
Before Starting The Workshop
Please ensure you have the latest version of R and RStudio installed on your machine. This is important, as some packages used in the workshop may not install correctly (or at all) if R is not up to date.
Data Set
We will begin with raw data, perform exploratory analyses, and learn how to plot results graphically. We’ll use a version of the Palmer Penguins dataset dataset from the Palmer Penguins package that we have adapted for the purposes of this course. The Palmer Penguins dataset provides morphological measurements (bill length, bill depth, flipper length, body mass) for three penguin species (Adélie, Chinstrap, and Gentoo) collected from three islands (Torgersen, Dream, and Biscoe) in the Palmer Archipelago, Antarctica.

Can you read the data into R? Can you plot relationship between bill length and flipper length? Can you calculate the mean body mass of penguins on the island of Dream? By the end of these lessons you will be able to explore key relationships in the data in minutes!
Introduction to RStudio
Throughout this lesson, we’re going to teach you some of the fundamentals of the R language as well as some best practices for organizing code for scientific projects that will make your life easier.
Basic layout
When you first open RStudio, you will be greeted by three panels:
- The interactive R console/Terminal (entire left)
- Environment/History/Connections (tabbed in upper right)
- Files/Plots/Packages/Help/Viewer (tabbed in lower right)
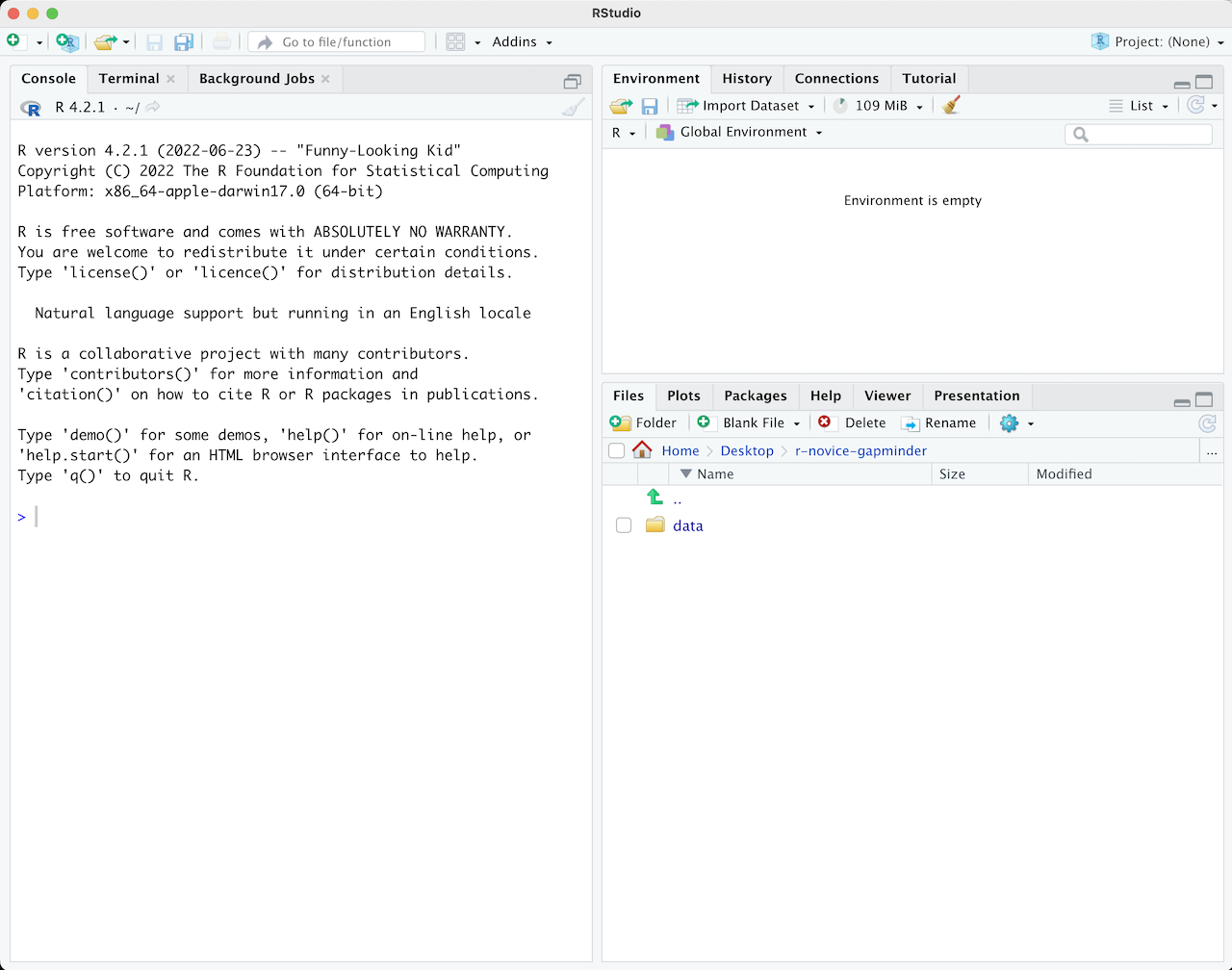
Once you open files, such as R scripts, an editor panel will also open in the top left.
Projects in R Studio
The scientific process is naturally incremental, and many projects start life as random notes, some code, then a manuscript, and eventually everything is a bit mixed together.
Managing your projects in a reproducible fashion doesn’t just make your science reproducible, it makes your life easier.
— Vince Buffalo (@vsbuffalo) April 15, 2013
Most people tend to organize their projects like this:
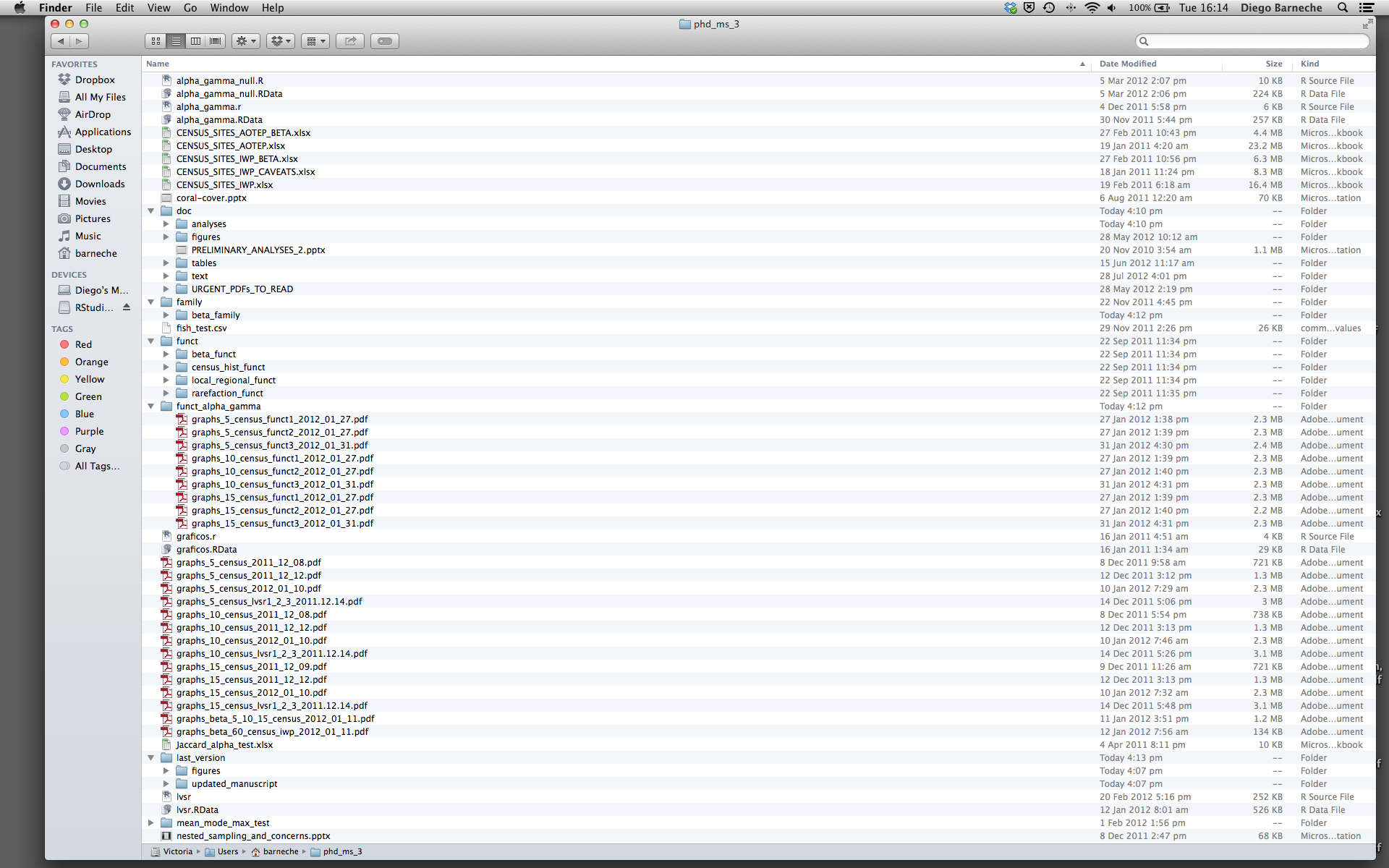
There are many reasons why we should ALWAYS avoid this:
- It is really hard to tell which version of your data is the original and which is the modified;
- It gets really messy because it mixes files with various extensions together;
- It probably takes you a lot of time to actually find things, and relate the correct figures to the exact code that has been used to generate it;
A good project layout will ultimately make your life easier:
- It will help ensure the integrity of your data;
- It makes it simpler to share your code with someone else (a lab-mate, collaborator, or supervisor);
- It allows you to easily upload your code with your manuscript submission;
- It makes it easier to pick the project back up after a break.
Creating a self-contained project
Fortunately, there are tools and packages which can help you manage your work effectively.
One of the most powerful and useful aspects of RStudio is its project management functionality. We’ll be using this today to create a self-contained, reproducible project.
We’re going to create a new project in RStudio:
- Click the “File” menu button, then “New Project”.
- Click “New Directory”.
- Click “New Project”.
- Type in the name of the directory to store your project, e.g. “my_project”.
- If available, select the checkbox for “Create a git repository.”
- Click the “Create Project” button.
Best practices for project organization
Although there is no “best” way to lay out a project, there are some general principles to adhere to that will make project management easier:
Treat data as read only
This is probably the most important goal of setting up a project. Data is typically time consuming and/or expensive to collect. Working with them interactively (e.g., in Excel) where they can be modified means you are never sure of where the data came from, or how it has been modified since collection. It is therefore a good idea to treat your data as “read-only”.
Data Cleaning
In many cases your data will be “dirty”: it will need significant preprocessing to get into a format R (or any other programming language) will find useful. This task is sometimes called “data munging”. Storing these scripts in a separate folder, and creating a second “read-only” data folder to hold the “cleaned” data sets can prevent confusion between the two sets.
Treat generated output as disposable
Anything generated by your scripts should be treated as disposable: it should all be able to be regenerated from your scripts.
There are lots of different ways to manage this output. Having an output folder with different sub-directories for each separate analysis makes it easier later. Since many analyses are exploratory and don’t end up being used in the final project, and some of the analyses get shared between projects.
Tip: Good Enough Practices for Scientific Computing
Good Enough Practices for Scientific Computing gives the following recommendations for project organization:
- Put each project in its own directory, which is named after the project.
- Put text documents associated with the project in the
docdirectory. - Put raw data and metadata in the
datadirectory, and files generated during cleanup and analysis in aresultsdirectory. - Put source for the project’s scripts and programs in the
srcdirectory, and programs brought in from elsewhere or compiled locally in thebindirectory. - Name all files to reflect their content or function.
Use Project Subfolders
Create data, src, and results
directories in your project directory.
Copy the files penguins_teaching.csv, weather-data.csv, and weather-data_v2.csv files from the zip file you downloaded as part of the lesson set up to the data/ folder within your project. We will load and use these files later in the course.
Tip: Working directory
Knowing R’s current working directory is important because when you need to access other files (for example, to import a data file), R will look for them relative to the current working directory.
Each time you create a new RStudio Project, it will create a new
directory for that project. When you open an existing
.Rproj file, it will open that project and set R’s working
directory to the folder that file is in.
You can check the current working directory with the
getwd() command, or by using the menus in RStudio.
- In the console, type
getwd()(“wd” is short for “working directory”) and hit Enter. - In the Files pane, double click on the
datafolder to open it (or navigate to any other folder you wish). To get the Files pane back to the current working directory, click “More” and then select “Go To Working Directory”.
You can change the working directory with setwd(), or by
using RStudio menus.
- In the console, type
setwd("data")and hit Enter. Typegetwd()and hit Enter to see the new working directory. - In the menus at the top of the RStudio window, click the “Session”
menu button, and then select “Set Working Directory” and then “Choose
Directory”. Next, in the windows navigator that opens, navigate back to
the project directory, and click “Open”. Note that a
setwdcommand will automatically appear in the console.
Workflow within RStudio
There are two main ways one can work within RStudio:
- Test and play within the interactive R console then copy code into a .R file to run later.
- This works well when doing small tests and initially starting off.
- It quickly becomes laborious
- Start writing in a .R file and use RStudio’s short cut keys for the Run command to push the current line, selected lines or modified lines to the interactive R console.
- This is a great way to start; all your code is saved for later
- You will be able to run the file you create from within RStudio or
using R’s
source()function.
In this lesson we’ll spend a lot of time working in the interactive console. We’ll move on to creating scripts in the next lesson.
Tip: Running segments of your code
RStudio offers you great flexibility in running code from within the editor window. There are buttons, menu choices, and keyboard shortcuts. To run the current line, you can
- click on the
Runbutton above the editor panel, or - select “Run Lines” from the “Code” menu, or
- hit Ctrl+Return in Windows or Linux or
⌘+Return on OS X. (This shortcut can also be seen
by hovering the mouse over the button). To run a block of code, select
it and then
Run. If you have modified a line of code within a block of code you have just run, there is no need to reselect the section andRun, you can use the next button along,Re-run the previous region. This will run the previous code block including the modifications you have made.
R scripts
Any commands that you write in the R console can be saved to a file
to be re-run again. Files containing R code to be run in this way are
called R scripts. R scripts have .R at the end of their
names to let you know what they are.
Introduction to R
Much of your time in R will be spent in the R interactive console.
This is where you will run all of your code, and can be a useful
environment to try out ideas before adding them to an R script file.
This console in RStudio is the same as the one you would get if you
typed in R in your command-line environment.
The first thing you will see in the R interactive session is a bunch of information, followed by a “>” and a blinking cursor. In many ways this is similar to the shell environment you learned about during the shell lessons: it operates on the same idea of a “Read, evaluate, print loop”: you type in commands, R tries to execute them, and then returns a result.
Using R as a calculator
The simplest thing you could do with R is to do arithmetic:
R
1 + 100
OUTPUT
[1] 101And R will print out the answer, with a preceding “[1]”. [1] is the index of the first element of the line being printed in the console.
If you type in an incomplete command, R will wait for you to complete it. If you are familiar with Unix Shell’s bash, you may recognize this behavior from bash.
OUTPUT
+Any time you hit return and the R session shows a “+” instead of a “>”, it means it’s waiting for you to complete the command. If you want to cancel a command you can hit Esc and RStudio will give you back the “>” prompt.
Tip: Canceling commands
If you’re using R from the command line instead of from within RStudio, you need to use Ctrl+C instead of Esc to cancel the command. This applies to Mac users as well!
Canceling a command isn’t only useful for killing incomplete commands: you can also use it to tell R to stop running code (for example if it’s taking much longer than you expect), or to get rid of the code you’re currently writing.
When using R as a calculator, the order of operations is the same as you would have learned back in school.
From highest to lowest precedence:
- Parentheses:
(,) - Exponents:
^or** - Multiply:
* - Divide:
/ - Add:
+ - Subtract:
-
R
3 + 5 * 2
OUTPUT
[1] 13Use parentheses to group operations in order to force the order of evaluation if it differs from the default, or to make clear what you intend.
R
(3 + 5) * 2
OUTPUT
[1] 16This can get unwieldy when not needed, but clarifies your intentions. Remember that others may later read your code.
R
(3 + (5 * (2 ^ 2))) # hard to read
3 + 5 * 2 ^ 2 # clear, if you remember the rules
3 + 5 * (2 ^ 2) # if you forget some rules, this might help
The text after each line of code is called a “comment”. Anything that
follows after the hash (or octothorpe) symbol # is ignored
by R when it executes code.
Really small or large numbers get a scientific notation:
R
2/10000
OUTPUT
[1] 2e-04Which is shorthand for “multiplied by 10^XX”. So
2e-4 is shorthand for 2 * 10^(-4).
You can write numbers in scientific notation too:
R
5e3 # Note the lack of minus here
OUTPUT
[1] 5000Mathematical functions
R has many built in mathematical functions. To call a function, we can type its name, followed by open and closing parentheses. Functions take arguments as inputs, anything we type inside the parentheses of a function is considered an argument. Depending on the function, the number of arguments can vary from none to multiple. For example:
R
getwd() #returns an absolute filepath
doesn’t require an argument, whereas for the next set of mathematical functions we will need to supply the function a value in order to compute the result.
R
sin(1) # trigonometry functions
OUTPUT
[1] 0.841471R
log(1) # natural logarithm
OUTPUT
[1] 0R
log10(10) # base-10 logarithm
OUTPUT
[1] 1R
exp(0.5) # e^(1/2)
OUTPUT
[1] 1.648721Don’t worry about trying to remember every function in R. You can look them up on Google, or if you can remember the start of the function’s name, use the tab completion in RStudio.
This is one advantage that RStudio has over R on its own, it has auto-completion abilities that allow you to more easily look up functions, their arguments, and the values that they take.
Typing a ? before the name of a command will open the
help page for that command. When using RStudio, this will open the
‘Help’ pane; if using R in the terminal, the help page will open in your
browser. The help page will include a detailed description of the
command and how it works. Scrolling to the bottom of the help page will
usually show a collection of code examples which illustrate command
usage.
Comparing things
We can also do comparisons in R:
R
1 == 1 # equality (note two equals signs, read as "is equal to")
OUTPUT
[1] TRUER
1 != 2 # inequality (read as "is not equal to")
OUTPUT
[1] TRUER
1 < 2 # less than
OUTPUT
[1] TRUER
1 <= 1 # less than or equal to
OUTPUT
[1] TRUER
1 > 0 # greater than
OUTPUT
[1] TRUER
1 >= -9 # greater than or equal to
OUTPUT
[1] TRUETip: Comparing Numbers
A word of warning about comparing numbers: you should never use
== to compare two numbers unless they are integers (a data
type which can specifically represent only whole numbers).
Computers may only represent decimal numbers with a certain degree of precision, so two numbers which look the same when printed out by R, may actually have different underlying representations and therefore be different by a small margin of error (called Machine numeric tolerance).
Instead you should use the all.equal function.
Further reading: http://floating-point-gui.de/
Variables and assignment
We can store values in variables using the assignment operator
<-, like this:
R
x <- 1/40
Notice that assignment does not print a value. Instead, we stored it
for later in something called a variable.
x now contains the value
0.025:
R
x
OUTPUT
[1] 0.025More precisely, the stored value is a decimal approximation of this fraction called a floating point number.
Look for the Environment tab in the top right panel of
RStudio, and you will see that x and its value have
appeared. Our variable x can be used in place of a number
in any calculation that expects a number:
R
log(x)
OUTPUT
[1] -3.688879Notice also that variables can be reassigned:
R
x <- 100
x used to contain the value 0.025 and now it has the
value 100.
Assignment values can contain the variable being assigned to:
R
x <- x + 1 #notice how RStudio updates its description of x on the top right tab
y <- x * 2
The right hand side of the assignment can be any valid R expression. The right hand side is fully evaluated before the assignment occurs.
Variable names can contain letters, numbers, underscores and periods but no spaces. They must start with a letter or a period followed by a letter (they cannot start with a number nor an underscore). Variables beginning with a period are hidden variables. Different people use different conventions for long variable names, these include
- periods.between.words
- underscores_between_words
- camelCaseToSeparateWords
What you use is up to you, but be consistent.
It is also possible to use the = operator for
assignment:
R
x = 1/40
But this is much less common among R users. The most important thing
is to be consistent with the operator you use. There
are occasionally places where it is less confusing to use
<- than =, and it is the most common symbol
used in the community. So the recommendation is to use
<-.
Vectorization
One final thing to be aware of is that R is vectorized, meaning that variables and functions can have vectors as values. A vector is 1 dimensional array of ordered values all of the same data type. For example:
R
1:5
OUTPUT
[1] 1 2 3 4 5We can assign a vector to a variable:
R
x <- 5:10
We can apply functions to all the elements of a vector:
R
(1:5) * 2
OUTPUT
[1] 2 4 6 8 10R
2^(1:5)
OUTPUT
[1] 2 4 8 16 32We can also create vectors “by hand” using the c()
function; this tersely named function is used to combine values
into a vector; these values can, themselves, be vectors:
R
c(2, 4, -1)
OUTPUT
[1] 2 4 -1R
c(x, 2, 2, 3)
OUTPUT
[1] 5 6 7 8 9 10 2 2 3Vectors aren’t limited to storing numbers:
R
c("a", "b", "c", "def")
OUTPUT
[1] "a" "b" "c" "def"R comes with a few built in constants, containing useful values:
R
LETTERS
letters
month.abb
month.name
We will use some of these in the examples that follow.
Vector lengths
We can calculate how many elements a vector contains using the
length() function:
R
length(x)
OUTPUT
[1] 6R
length(letters)
OUTPUT
[1] 26Subsetting vectors
Having defined a vector, it’s often useful to extract parts
of a vector. We do this with the [] operator. Using the
built in month.name vector:
R
month.name[2]
OUTPUT
[1] "February"R
month.name[2:4]
OUTPUT
[1] "February" "March" "April" Let’s unpick the second example; 2:4 generates the
sequence 2,3,4. This gets passed to the extract operator
[]. We can also generate this sequence using the
c() function:
R
month.name[c(2,3,4)]
OUTPUT
[1] "February" "March" "April" Vector numbering in R starts at 1
In many programming languages (C and python, for example), the first element of a vector has an index of 0. In R, the first element is 1.
Values are returned in the order that we specify the indices. We can extract the same element more than once:
R
month.name[4:2]
OUTPUT
[1] "April" "March" "February"R
month.name[c(1,1,2,3,4)]
OUTPUT
[1] "January" "January" "February" "March" "April" If we try and extract an element that doesn’t exist in the vector,
the missing values are NA:
R
month.name[10:13]
OUTPUT
[1] "October" "November" "December" NA Missing data
NA is a special value, that is used to represent “not
available”, or “missing”. If we perform computations which include
NA, the result is usually NA:
R
1 + NA
OUTPUT
[1] NAThis raises an interesting point; how do we test if a value is
NA? This doesn’t work:
R
x <- NA
x == NA
OUTPUT
[1] NAHandling special values
There are a number of special functions you can use to handle missing data, and other special values:
-
is.nawill return all positions in a vector, matrix, or data.frame containingNA. - likewise,
is.nan, andis.infinitewill do the same forNaNandInf. -
is.finitewill return all positions in a vector, matrix, or data.frame that do not containNA,NaNorInf. -
na.omitwill filter out all missing values from a vector
Skipping and removing elements
If we use a negative number as the index of a vector, R will return every element except for the one specified:
R
month.name[-2]
OUTPUT
[1] "January" "March" "April" "May" "June" "July"
[7] "August" "September" "October" "November" "December" We can skip multiple elements:
R
month.name[c(-1, -5)] # or month.name[-c(1,5)]
OUTPUT
[1] "February" "March" "April" "June" "July" "August"
[7] "September" "October" "November" "December" Tip: Order of operations
A common error occurs when trying to skip slices of a vector. Most people first try to negate a sequence like so:
R
month.name[-1:3]
This gives a somewhat cryptic error:
ERROR
Error in month.name[-1:3]: only 0's may be mixed with negative subscriptsBut remember the order of operations. : is really a
function, so what happens is it takes its first argument as -1, and
second as 3, so generates the sequence of numbers:
-1, 0, 1, 2, 3.
The correct solution is to wrap that function call in brackets, so
that the - operator is applied to the sequence:
R
-(1:3)
OUTPUT
[1] -1 -2 -3R
month.name[-(1:3)]
OUTPUT
[1] "April" "May" "June" "July" "August" "September"
[7] "October" "November" "December" Subsetting with logical vectors
As well as providing a list of indices we want to keep (or delete, if
we prefix them with -), we can pass a logical
vector to R indicating the indices we wish to select:
R
fourmonths <- month.name[1:4]
fourmonths
OUTPUT
[1] "January" "February" "March" "April" R
fourmonths[c(TRUE, FALSE, TRUE, TRUE)]
OUTPUT
[1] "January" "March" "April" What happens if we supply a logical vector that is shorter than the vector we’re extracting the elements from?
R
fourmonths[c(TRUE,FALSE)]
OUTPUT
[1] "January" "March" This illustrates the idea of vector recycling; the
[] extract operator “recycles” the subsetting vector:
R
fourmonths[c(TRUE,FALSE,TRUE,FALSE)]
OUTPUT
[1] "January" "March" This can be useful, but can easily catch you out.
The idea of selecting elements of a vector using a logical subsetting vector may seem a bit esoteric, and a lot more typing than just selecting the elements you want by index. It becomes really useful when we write code to generate the logical vector:
R
my_vector <- c(0.01, 0.69, 0.51, 0.39)
my_vector > 0.5
OUTPUT
[1] FALSE TRUE TRUE FALSER
my_vector[my_vector > 0.5]
OUTPUT
[1] 0.69 0.51Tip: Combining logical conditions
There are many situations in which you will wish to combine multiple logical criteria. For example, we might want to find all the elements that are between two values. Several operations for combining logical vectors exist in R:
-
&, the “logical AND” operator: returnsTRUEif both the left and right areTRUE. -
|, the “logical OR” operator: returnsTRUE, if either the left or right (or both) areTRUE.
The recycling rule applies with both of these, so
TRUE & c(TRUE, FALSE, TRUE) will compare the first
TRUE on the left of the & sign with each
of the three conditions on the right.
You may sometimes see && and ||
instead of & and |. These operators do not
use the recycling rule: they only look at the first element of each
vector and ignore the remaining elements. The longer operators are
mainly used in programming, rather than data analysis.
-
!, the “logical NOT” operator: convertsTRUEtoFALSEandFALSEtoTRUE. It can negate a single logical condition (e.g.!TRUEbecomesFALSE), or a whole vector of conditions(e.g.!c(TRUE, FALSE)becomesc(FALSE, TRUE)).
Additionally, you can compare the elements within a single vector
using the all function (which returns TRUE if
every element of the vector is TRUE) and the
any function (which returns TRUE if one or
more elements of the vector are TRUE).
Challenge 2
What will be the value of each variable after each statement in the following program?
R
mass <- 47.5
depth <- 122
mass <- mass * 2.3
depth <- depth - 20
R
mass <- 47.5
This will give a value of 47.5 for the variable mass
R
depth <- 122
This will give a value of 122 for the variable depth
R
mass <- mass * 2.3
This will multiply the existing value of 47.5 by 2.3 to give a new value of 109.25 to the variable mass.
R
depth <- depth - 20
This will subtract 20 from the existing value of 122 to give a new value of 102 to the variable depth.
Challenge 3
Run the code from the previous challenge, and write a command to compare mass to depth. Is mass larger than depth?
One way of answering this question in R is to use the
> to set up the following:
R
mass > depth
OUTPUT
[1] TRUEThis should yield a boolean value of TRUE since 109.25 is greater than 102.
Challenge 4
Return a vector containing the letters of the alphabet in reverse order
We can extract the elements in reverse order by generating the
sequence 26, 25, … 1 using the : operator:
R
letters[26:1]
OUTPUT
[1] "z" "y" "x" "w" "v" "u" "t" "s" "r" "q" "p" "o" "n" "m" "l" "k" "j" "i" "h"
[20] "g" "f" "e" "d" "c" "b" "a"Although this works, it makes the assumption that
letters will always be length 26. Although this is probably
a safe assumption in English, other languages may have more (or fewer)
letters in their alphabet. It is good practice to avoid assuming
anything about your data.
We can avoid assuming letters is length 26, by using the
length() function:
R
letters[length(letters):1]
OUTPUT
[1] "z" "y" "x" "w" "v" "u" "t" "s" "r" "q" "p" "o" "n" "m" "l" "k" "j" "i" "h"
[20] "g" "f" "e" "d" "c" "b" "a"Challenge 5
Given the following code:
R
x <- c(5.4, 6.2, 7.1, 7.5, 4.8)
x
OUTPUT
[1] 5.4 6.2 7.1 7.5 4.8Come up with at least 3 different commands that will produce the following output:
OUTPUT
[1] 6.2 7.1 7.5After you find 3 different commands, compare notes with your neighbour. Did you have different strategies?
R
x[2:4]
OUTPUT
[1] 6.2 7.1 7.5R
x[-c(1,5)]
OUTPUT
[1] 6.2 7.1 7.5R
x[c(2,3,4)]
OUTPUT
[1] 6.2 7.1 7.5R
x[c(FALSE, TRUE, TRUE, TRUE, FALSE)]
OUTPUT
[1] 6.2 7.1 7.5(We can use vector recycling to make the previous example slightly shorter:
R
x[c(FALSE, TRUE, TRUE, TRUE)]
OUTPUT
[1] 6.2 7.1 7.5The first element of the logical vector will be recycled)
We can construct a logical test to generate the logical vector:
R
x[x > 6]
OUTPUT
[1] 6.2 7.1 7.5Unpicking this example, x > 6 generates the logical
vector:
OUTPUT
[1] FALSE TRUE TRUE TRUE FALSEWhich is passed to the [ ] subsetting operator.
Further reading
We recommend the following resources for some additional reading on the topic of this episode:
Key Points
- Use RStudio to write and run R programs.
- R has the usual arithmetic operators and mathematical functions.
- Use
<-to assign values to variables. - Use RStudio to create and manage projects with a consistent structure.
- Treat raw data as read-only.
- Treat generated output as disposable.
